Community Data Challenges
Data challenges are competitions where participants solve problems using open-source datasets, providing community members a way to directly interact with data generating labs. The Allen Institute has spearheaded multiple data challenges targeting distinct scientific challenges in brain cell typing.

SEA-AD DREAM Challenge (active)
Alzheimer's disease is a common neurodegenerative disorder characterized by progressive cognitive decline and the accumulation of amyloid-beta plaques and Tau tangles in the brain. Recent advances in single-cell transcriptomics have provided unprecedented insights into the cellular composition of the aging and diseased brain, generating rich, high-dimensional datasets that may reveal molecular signatures of vulnerability and resilience. This crowdsourced challenge aims to link these two data modalities, calling on the research community to develop models that predict Alzheimer's neuropathological burden from single-cell gene expression data. Challenge closes on October 22nd, with ongoing submissions allowed until then.
Link to challenge | Publication (Gabitto, Travaglini, et al 2024)
MapMySections (completed)
Transgenic lines and viral tools are invaluable for targeting specific cell populations in the brains of mice and other species. The recent identification of over 5,000 distinct mouse brain cell types, each with a unique gene expression profile and spatial location, presents new opportunities. Not only can researchers develop novel cell-type-specific genetic tools, but they can also better characterize the specificity of thousands of existing tools already in use in neuroscience. Precisely defining this connection is crucial for correctly interpreting the results of experiments that use these tools. While a detailed characterization can be achieved through costly and time-consuming methods like single-cell RNA sequencing and cell sorting, a more direct approach is needed. This challenge tasked participants with creating an algorithm that can accurately infer cell types directly from fluorescent images. Focused on the primary visual cortex (VISp), the goal was to develop a tool that matches fluorescent images of genetic tools to their most likely spatial transcriptomic cell types. This data challenge closed in May 2025. Challenge details, tools and algorithms submitted by all entrants, and challenge solutions are all available at the data challenge website.
Link to challenge | Webinar | Allen Brain Cell Atlas | Genetics tools atlas | Publication (Ben-Simon, Hooper, Narayan, Daigle, et al 2025)
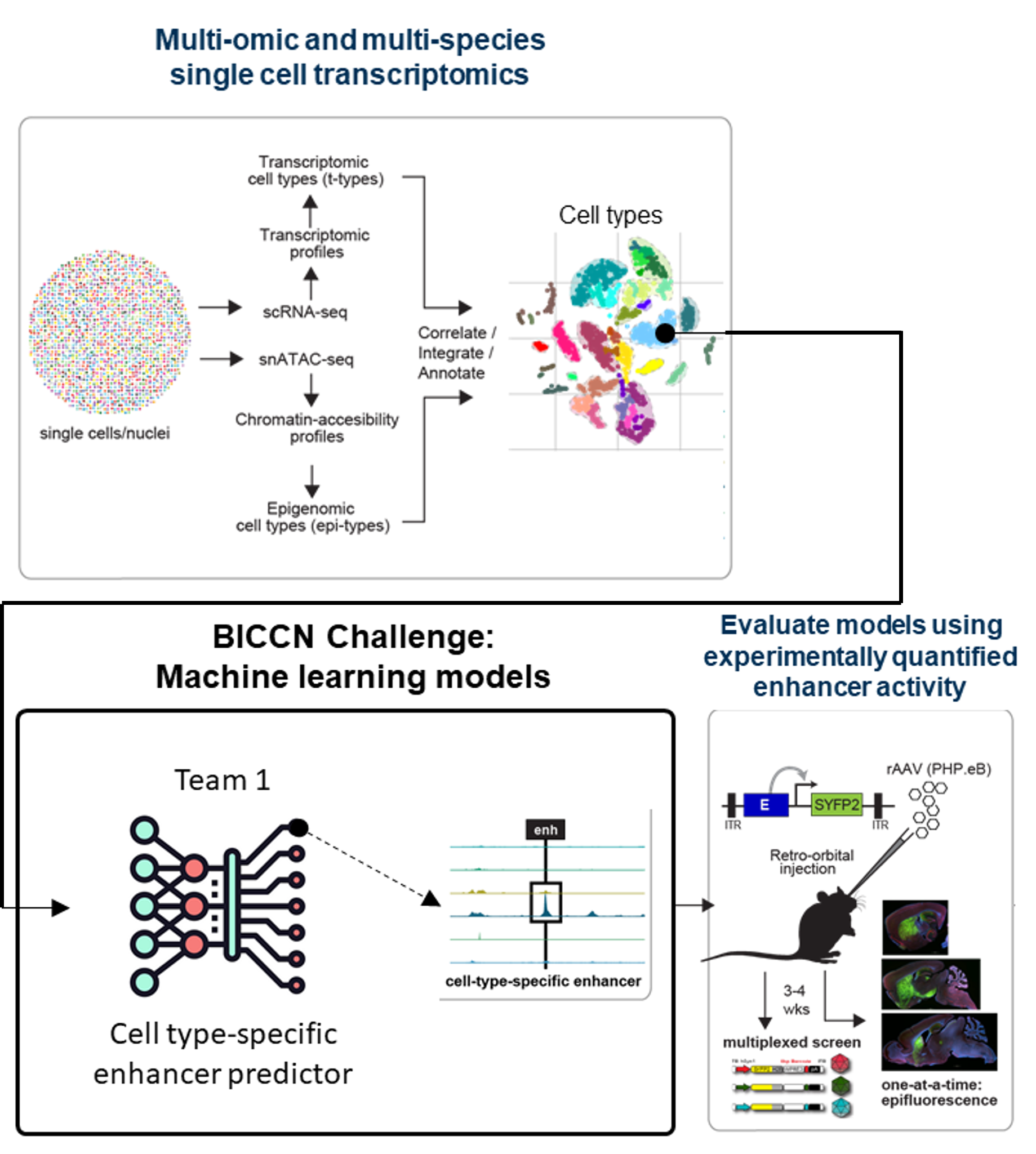
BICCN Enhancer Challenge (completed; active community benchmark)
Single-cell sequencing technologies enable molecular profiling of millions of cells to define atlases of cell types across tissues and species. Recent advancements enable measurement of multiple genomic features (e.g. RNA and open chromatin) from the same cell. Multi-omic profiling of the brain across species enables high-resolution alignment of ‘homologous’ cell types that have conserved and specialized molecular features. In this competition, teams predicted cell type-specific enhancers using new multi-omics and multi-species atlases of cell types in the primary motor cortex. Teams were evaluated against in vivo activity of enhancers in the mouse brain from a collection of several hundred enhancers that were experimentally screened by Ben-Simon et al. 2024. Progress will serve as a foundation for targeted exploration of cell types in brain circuitry across species. Challenge details and leaderboard are available at the data challenge website along with guidelines for continued use by the community as a benchmark.
Link to challenge | Genetics tools atlas | Publication (Johansen, Kempynck, et al 2025)
MapMySpikes (completed)
Currently [circa 2024], a major challenge for neuroscientists is the inability to easily map electrophysiological data—such as that from patch-clamp recordings—to specific cell types. This disconnect makes it difficult for researchers to interpret their findings and integrate them with the vast transcriptomic knowledge of the Allen Brain Map. To bridge this gap, a data challenge was launched, tasking participants with developing an algorithm that accurately matches electrophysiological data to known transcriptomic cell types in the mouse primary visual and motor cortices and present these results in a user-friendly tool. This data challenge closed in June 2024. Challenge details, a subset of tools and algorithms submitted by entrants, and challenge solutions are all available at the data challenge website.
Link to challenge | Webinar | Mouse Patch-seq VISp | Cell Type Knowledge Explore
Your browser is out-of-date!
Update your browser to view this website correctly.